Page 199 - Kaleidoscope Academic Conference Proceedings 2024
P. 199
Innovation and Digital Transformation for a Sustainable World
machine learning algorithm for dimensionality reduction,
particularly useful for visualizing high-dimensional data. In
our context, t-SNE helps to reduce the high-dimensional word
embeddings to a 2D space, allowing us to visualize and
analyze the relationships between different oncology terms.
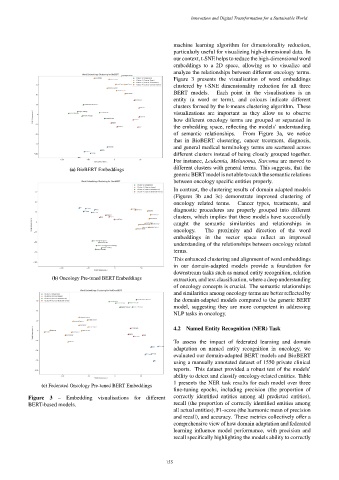
Figure 3 presents the visualisation of word embeddings
clustered by t-SNE dimensionality reduction for all three
BERT models. Each point in the visualisations is an
entity (a word or term), and colours indicate different
clusters formed by the k-means clustering algorithm. These
visualizations are important as they allow us to observe
how different oncology terms are grouped or separated in
the embedding space, reflecting the models’ understanding
of semantic relationships. From Figure 3a, we notice
that in BioBERT clustering, cancer treatment, diagnosis,
and general medical terminology terms are scattered across
different clusters instead of being closely grouped together.
For instance, Leukemia, Melanoma, Sarcoma are moved to
different clusters with general terms. This suggests, that the
(a) BioBERT Embeddings
generic BERTmodel isnotable tocatch thesemanticrelations
between oncology specific entities properly.
In contrast, the clustering results of domain adapted models
(Figures 3b and 3c) demonstrate improved clustering of
oncology related terms. Cancer types, treatments, and
diagnostic procedures are properly grouped into different
clusters, which implies that these models have successfully
caught the semantic similarities and relationships in
oncology. The proximity and direction of the word
embeddings in the vector space reflect an improved
understanding of the relationships between oncology related
terms.
This enhanced clustering and alignment of word embeddings
in our domain-adapted models provide a foundation for
downstream tasks such as named entity recognition, relation
(b) Oncology Pre-tuned BERT Embeddings extraction, and text classification, where a deep understanding
of oncology concepts is crucial. The semantic relationships
and similarities among oncology terms are better reflected by
the domain-adapted models compared to the generic BERT
model, suggesting they are more competent in addressing
NLP tasks in oncology.
4.2 Named Entity Recognition (NER) Task
To assess the impact of federated learning and domain
adaptation on named entity recognition in oncology, we
evaluated our domain-adapted BERT models and BioBERT
using a manually annotated dataset of 1550 private clinical
reports. This dataset provided a robust test of the models’
ability to detect and classify oncology-related entities. Table
1 presents the NER task results for each model over three
(c) Federated Oncology Pre-tuned BERT Embeddings
fine-tuning epochs, including precision (the proportion of
Figure 3 – Embedding visualisations for different correctly identified entities among all predicted entities),
BERT-based models. recall (the proportion of correctly identified entities among
all actual entities), F1-score (the harmonic mean of precision
and recall), and accuracy. These metrics collectively offer a
comprehensive view of how domain adaptation and federated
learning influence model performance, with precision and
recall specifically highlighting the models ability to correctly
– 155 –